Post processing of a very large Mustguseal alignment using MAPU
Large alignment of proteins within a superfamily incorporates all currently known variability among a diverse set of evolutionary distantly related homologues and thus can help at studying the sequence/ structure/ function relationship, but can be very hard to further process due to its huge dimensions. This page introduces a software - the Mustguseal Alignment Postprocess Utility (MAPU) - to extract only the most informative part of an alignment to facilitate its further analysis.
Overview
Mustguseal can automatically construct very large alignments of protein families/superfamilies that include thousands of proteins, tens of thousands of alignment columns (due an extremely high amount of gaps as a result of sequence and structural variability among evolutionary distantly related homologues), and hundreds of thousands lines of text. The Mustguseal Alignment Postprocess Utility (MAPU) is a Java-dependent software intended to postprocess (trim) very large alignments of protein superfamilies created by Mustguseal. The purpose of MAPU is to remove the least informative columns and thus significantly reduce the size of the alignment and computational resources/time required for its further processing/analysis (e.g., by Zebra/pocketZebra/visualCMAT methods).
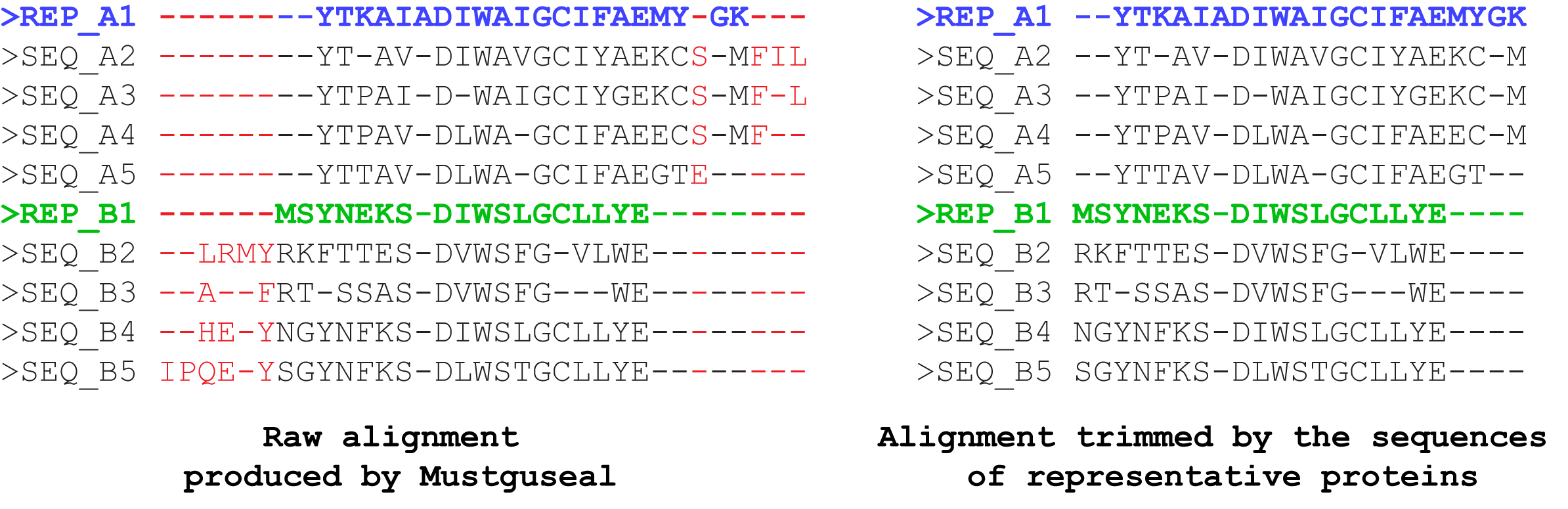
The MAPU reads an alignment of proteins sequences/structures in FASTA format (e.g., the final alignment produced by Mustguseal) and removes columns that:
- contain more than T% of gaps (i.e., are overpopulated by gaps and thus are not informative);
- AND
- are not represented by any of the user-defined reference proteins (i.e., contain a gap in all corresponding sequences).
I.e., only the alignment columns that are represented by at least one user-defined reference protein OR contain at most T% of gaps will be printed to the output file, and all other columns will be dismissed. The trimming will not lead to loss of information because further analysis of the final alignment is usually focused only on the columns which have a low content of gaps and contain amino acid residues of the representative proteins, and this information will be identical in the original and trimmed versions of the alignment. The purpose of trimming is to decrease the size of the final alignment file which can be crucial when dealing with large sets of diverse sequences and structures. E.g., the untrimmed version of the final alignment may occupy several hundred megabytes of the disk space due to a high content of gaps, and trimming can decrease the size by ~98% percent to just a few megabytes, which is very convenient for further analysis.
The input to MAPU is (1) an alignment file in FASTA format, (2) the full names of one or more reference proteins, and (3) the upper threshold for gaps for a column to be considered informative. Select the reference protein based on your particular task and primary interest. It can be the target protein selected for the further experimental design, the most studied member of the superfamily, or a protein which you are the most familiar with. Select T based on the requirements of the software which will be further used for analysis of the created alignment, but usually within 5-30%.
The output from MAPU is an alignment file in FASTA format. While MAPU is expected to preserve the most valuable columns of your alignment and should be safe to use in most cases it can, in principle, result in loss of information. Use common sense.
The download section
Download the MAPU v. 1.1 2018-03-12: [download]
Download the example data: [download]
Example
We further discuss the capabilities of MAPU on a particular example.
- Download the example data using the link above and extract files from the archive;
- The
FINAL.fastafile corresponds to an alignment of protein kinases. It contains 5136 proteins, 27695 alignment columns, 2383104 lines of text, and is worth 139MB of disc space. Further visual or automated analysis of such a large file can be problematic;
- For this example we will select the 1R3C PDB protein as the reference (this protein was selected for further study in a particular project of ours). To learn the exact name of the protein in the alignment run this command:
cat FINAL.fasta | grep '>' | nl | grep -i 1r3c 4675
>1r3c_A
- Run the MAPU to preserve only alignment columns which contain the reference protein 1R3C OR contain not more than 50% of gaps, which are assumed to be the most content-rich:
java -jar mustguseal_postprocess.jar -f FINAL.fasta -p 1r3c_A -g 50 -o FINAL_MAPU.fasta
The processing should take ~10 seconds on a modern Desktop computer. To process larger alignments you may need to increase the memory consumption for the Java machine:
java -jar -Xms256m -Xmx16384m mustguseal_postprocess.jar -f FINAL.fasta -p 1r3c_A -g 50 -o FINAL_MAPU.fasta
- The following interactive output should be produced by MAPU:
Input: The input multiple alignment file is FINAL.fasta
Input: The input multiple alignment file is FINAL.fasta
Input: The output file name is FINAL_MAPU.fasta
Input: The reference protein is 1r3c_A
Input: The maximum allowed gap occurrence in a column is 50.0%
Info: Reading MSA from file FINAL.fasta
Info: Input MSA's dimensions are proteins=5136 columns=27695
Info: Calculating gap occurrence in the input alignment
Info: Selecting alignment columns which contain at most 50.0% of gaps
Info: In total 327 alignment columns contain at most 50.0% of gaps (i.e., 27368 columns contain more gaps)
Info: Searching for the reference protein entitled 1r3c_A
Info: Protein entitled 1r3c_A contains 366 amino acid residues
Info: Protein entitled 1r3c_A contains 42 columns which contain more than 50.0% of gaps
Info: In total 3 alignment columns contain at most 50.0% of gaps and are not represented by the reference protein
Info: Printing columns which contain at least one amino acid residue for the reference protein OR contain at most 50.0% of gaps
Info: Output MSA's dimensions are proteins=5136 columns=369 Done!
- The output file
FINAL_MAPU.fastacontains just 369 the most content-rich alignment columns and is worth only 2.4MB of the disk space. Thus, MAPU was able to shrink the alignment by 98.3% by removing the least informative columns;
- The alignment file
FINAL_MAPU.fastaand PDB file of the representative protein 1R3C can be further submitted to Zebra/pocketZebra/visualCMAT web-server for analysis.